PMI Project Aims
The goal of the Plant-Microbe Interfaces SFA is to gain a deeper understanding of the diversity and functioning of mutually beneficial interactions between plants and microbes in the rhizosphere. The plant-microbe interface is the boundary across which a plant senses, interacts with, and may alter its associated biotic and abiotic environments. Understanding the exchange of energy, information, and materials across the plant-microbe interface at diverse spatial and temporal scales is our ultimate objective. Our ongoing efforts focus on characterizing and interpreting such interfaces using systems comprising plants and microbes, in particular the poplar tree (Populus) and its microbial community in the context of favorable plant microbe interactions. We seek to define the relationships among these organisms in natural settings, dissect the molecular signals and gene-level responses of the organisms using natural and model systems, and interpret this information using advanced computational tools.
The advances anticipated here will set the stage for detailed understanding of other symbiotic relationships and of natural routes to ecosystem response to climate change, the cycling and sequestration of carbon in terrestrial environments, and the development and management of renewable energy sources.
Importance of plant-microbe interfaces
The beneficial association of plants and microbes exemplifies a complex, multiorganismal system that is shaped by the participating organisms and the environmental forces acting on them. Often these plant-microbe interactions can benefit plant productivity and performance by, for example, (1) affecting nutrient uptake and growth allocation, (2) influencing plant hormone signaling, (3) inducing catabolism of toxic compounds, and (4) conferring resistance to pathogens. In both natural and engineered systems, plants and microbes function collectively to determine the responses of terrestrial ecosystems to global changes as well as to offer potential as dedicated feedstocks in a renewable, carbon-neutral economy.
Populus
Populus, is a dominant perennial component of temperate forests, has the broadest geographic distribution of any North American tree genus and is the model woody perennial organism. Populus was chosen as the first tree genome to be sequenced, and numerous tools for manipulating its genetics and physiology are available. Further, Populus is a leading candidate for bioenergy production and provides researchers with ecosystem-scale insights into the central role of plants in carbon sequestration and cycling in terrestrial ecosystems. As a perennial woody plant, Populus also is among only a few plant species that host both endo- and ectomycorrhizal fungal associates. Numerous other types of microorganisms can be found within, or closely associated with, various Populus tissues, and these organisms may range from highly beneficial to pathogenic with respect to host fitness. Ultimately, an improved fundamental understanding of plant-microbe interfaces will enable the use of indigenous or engineered systems to address challenges as diverse as bioenergy production, environmental remediation, and carbon cycling and sequestration. Specifically, elucidating the mechanisms involved in energy, information, and material transduction across interfaces will be critical for interpreting environmental responses based on genomic signatures. Three interrelated scientific aims create integrating themes for this SFA and drive needed advances in analytical and computational technologies.
Objective 1: Defining the molecular determinants for selection and maintenance of mutualistic relationships between Populus and specific members of its microbiome
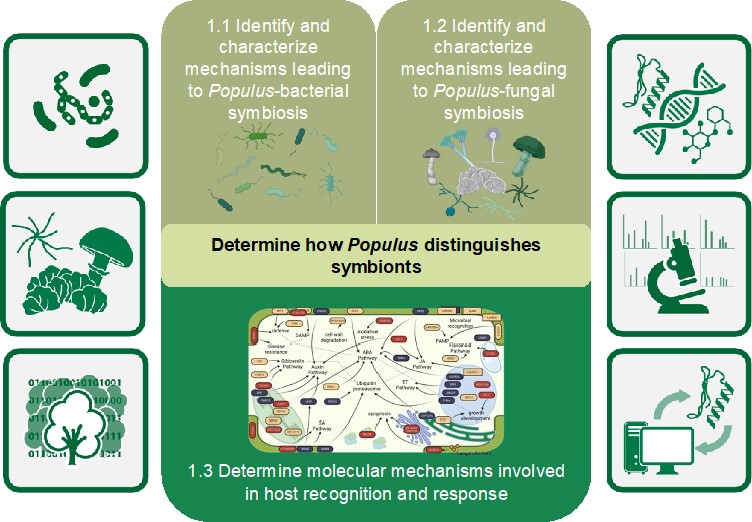
Our first objective seeks to define the molecular determinants for selection and maintenance of mutualistic relationships between Populus and specific members of its microbiome. This objective addresses the question of how the host selects specific microbial partners from a biologically diverse environment. We hypothesize that specific signaling systems, cell surface receptors, and downstream signaling events lead to general responses in Populus that distinguishes symbionts along a mutualism-commensalism-parasitism continuum. Previous PMI project discoveries defined specific genetic determinants, in the host and microbe, that lead to symbiotic associations. Furthermore, advanced computational tools have been developed to extract these features from the multiple, diverse layers of “omics” (i.e., genomics, transcriptomics, proteomics, and metabolomics) information. These advances are being used for expanding our studies to microbes that span the symbiotic spectrum, from mutualist to commensal to pathogen, to unlock the mechanisms by which Populus allows specific interactions. Using our plant and microbial resources, we are determining the commonalities and differences in host perception and response to diverse members of Populus’ microbial community to determine how Populus distinguishes “friend” from “foe.”
Objective 2: Deconstructing the chemical, physical and biological complexity of plant-microbial communities
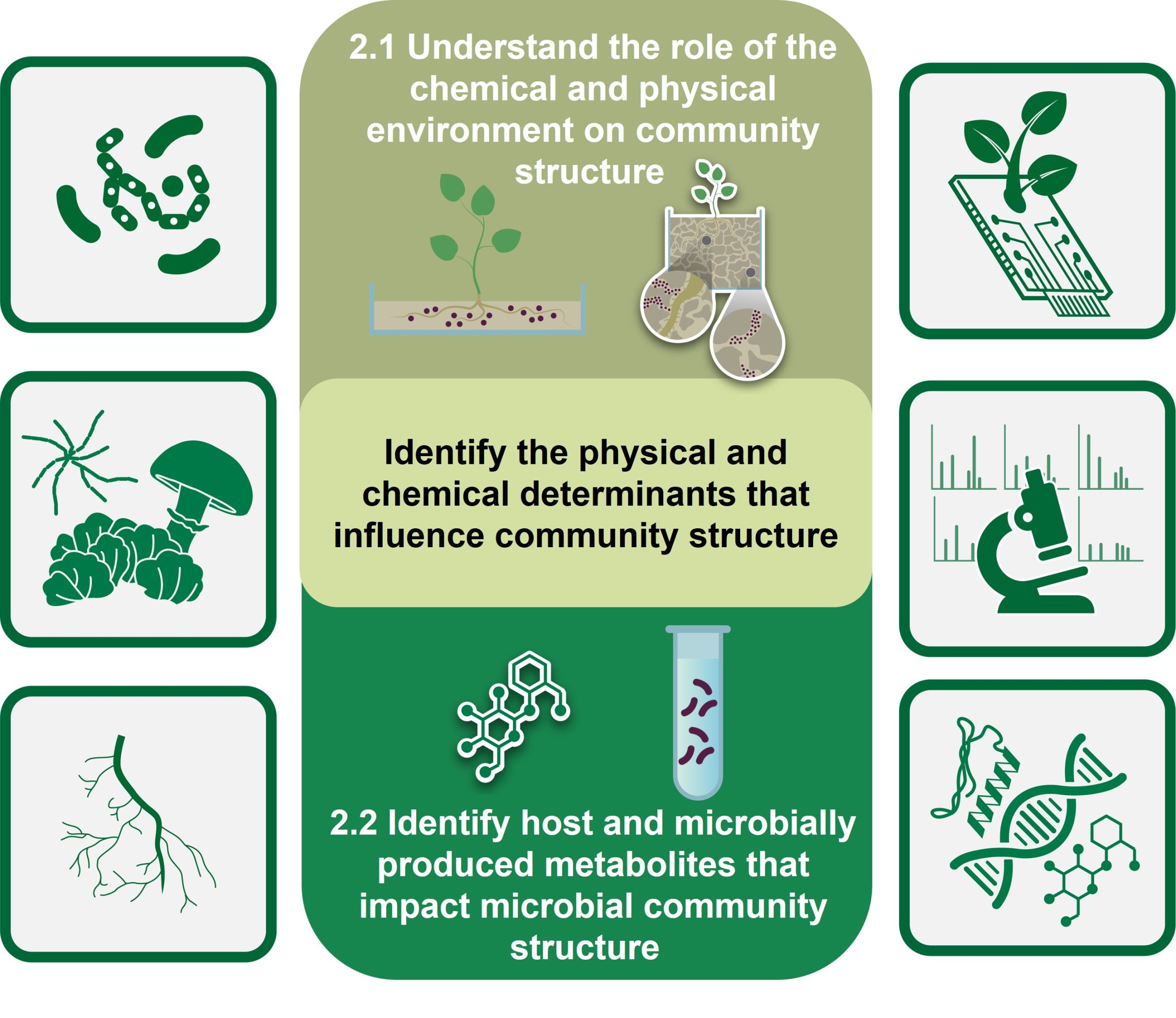
Our second objective examines how physical and chemical heterogeneity in the rhizosphere structures microbial community organization and function. This objective focuses on understanding how local microenvironments in the rhizosphere, the surrounding region under influence of the root, impact and are impacted by bacterial and fungal abundance and distribution. These efforts build off our successes in creating microscale habitats and in integrating chemical and physical imaging. We are visualizing and quantifying how local variations in the physical and chemical environment around the plant root impact recruitment and assembly of microbial communities and investigating the roles of specialized metabolite production in these processes. We hypothesize that the rhizosphere microbiome is structured in a way that reduces compositional complexity, and that this complexity is further reduced due to the shared functional capabilities of community members. Determining the spatial and temporal dynamics of local microenvironments and their role in structuring Populus’ microbiome are being accomplished using omics, imaging, and analytical methodologies. We expect that these studies will reveal new metabolites, advance methods for monitoring complex communities, and provide fundamental insights into how the microbiome is structured and proportioned in response to local environmental perturbations.
Objective 3: Modulation of host stress tolerance by the microbiome
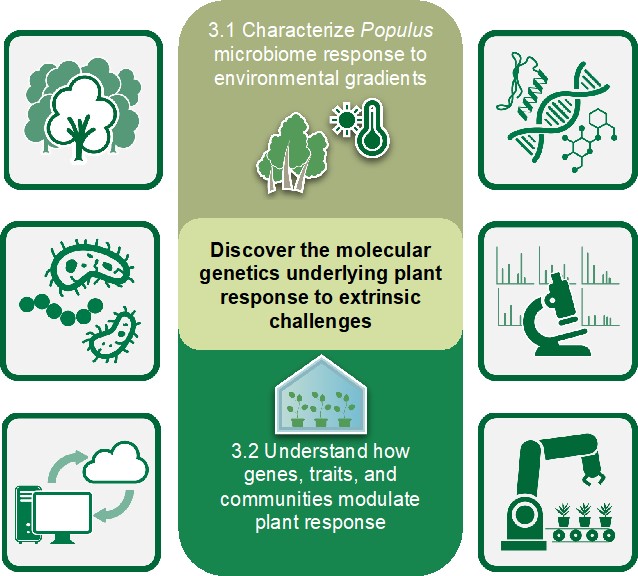
Our third objective evaluates the role of the Populus microbiome in the host’s ability to respond to environmental stressors. Our recent work demonstrates that the Populus microbiome is shaped by host genetic and environmental interactions that change through time. Furthermore, we have shown that warming adapted microbiomes can transmit thermotolerance to axenic laboratory grown plants. However, it is unclear which microbiome members and what functional traits of the microbial communities are responsible for the beneficial effects on the plant host. Within this objective, we hypothesize that microbial community composition modulates the host stress response, such that stress prone environments select for organisms and traits that complement and/or enhance host response. Through an integrated combination of natural system studies, multi-omic techniques, microbiome transfer experiments, defined constructed community approaches, genetic manipulation of host-microbe composition, and advanced computational tools, this third objective seeks to disentangle distinguishing traits, relationships, and mechanisms within the plant-microbe interface that influence host stress tolerance. Key organisms or traits identified that are important for host stress tolerance will feed back into new hypotheses and research directions within Objectives 1 and 2.


