Plant Microbe-Interfaces Objective 1
Understanding the progression of molecular events involved in selecting and maintaining a mutualistic relationship between Populus and specific members of its microbiome.
In this objective, we will focus on revealing the progression of molecular level events involved in selecting specific host-microbe relationships. The molecular events leading to bacterial-hit recognition and establishment of endophytic bacteria are not well understood. In formation on bacterial localization is lacking and characteristic physical structures, present in other microbial associations are not easily recognized. PMI efforts have defined numerous bacterial isolates that show niche specificity and are capable of being genetically manipulated. Using comparative genomics, and a parallelized taxonomic assignment algorithm (ParaKraken), which has been developed for analyzing meta-comics data, we are detecting genes specific to colonization and microbiome taxa that further depend on host genotype and environmental conditions. Leveraging our computational tools, -omics based analyses, and our extensive resource of cultured isolates, we are identifying the gene and bacterial and/or fungal hubs that are critical to how Populus selects its microbial symbionts.
Objective 1 Tasks
- Task 1.1 – Defining the molecular determinants for Populus-bacterial recognition and establishment.
- The focus will be on an in-depth study of PtLecRLK1-based signal transduction.
- Task 1.2 – Defining the molecular determinants for Populus-fungal interactions.
- The focus will be on the genes responsible for selective fungal recognition and colonization.
- Task 1.3 – Computationally based elucidation of the molecular determinants of diverse Populus-microbe interactions.
- We will use refined, model Populus-Bacterial systems, along with computational analyses, to identify the genes and molecules that underlie niche specific, mutualistic associations.
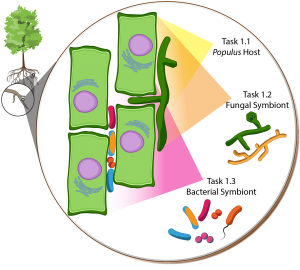
Objective 1 overview. Selective Populus-microbe interactions will be examined from the host perspective (Task 1.1), fungal perspective (Task 1.2), and bacterial perspective (Task 1.3).


